Species- and Site-Specific Genome Editing in Complex Bacterial Communities A novel toolset and approach to modify bacterial genomes within complex laboratory consortia.
Rubin, BE; Diamond, S; Cress, BF; Crits-Christoph A, Shivram H, He C, Xu M, Zhou Z, Smith SJ, Smock DC, Tang K, Owens TK, Krishnappa N, Sachdeva R, Barrangou R, Deutschbauer AM, Banfield JF, Doudna JA. (2021) Targeted Genome Editing of Bacteria Within Microbial Communities. Nature Microbiology 2021 [DOI]: 10.1038/s41564-021-01014-7

Most bacteria live together in complex communities like those in and around plant roots or those in the human gut and cannot be isolated in a lab. The contributions of individual microbes in these communities result in collective behaviors, and tools to dissect microbiomes at a community level are very limited. Typically, to study the function of of a bacterium, or its genes, scientists currently need to isolate individual species to enable them to alter or remove a gene in the bacterium’s genome to see how that modification changes the behavior of the bacterium. By developing and combining two new experimental tools (ET-Seq and DART) m-CAFEs researchers at University of California Berkeley in collaboration with North Carolina State University and Lawrence Berkeley National Lab are able to specifically modify the DNA of individual bacteria while they exist in a community as well as track these modifications as the community grows, which can help examine the functions of genes within uncultivated organisms. These were first applied to consortia of organisms mimicking these soil microbiomes; subsequent applications of the technique demonstrated the utility across diverse hosts and communities. Critically, this same approach can help examine the interactions of bacteria by enabling scientists to study how each member (and its genes) contributes to the community.
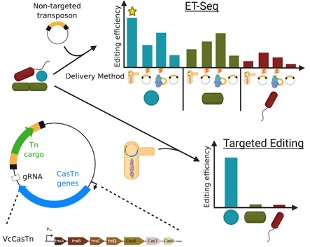
Methods developed for targeted genome editing in microbial communities: ET-Seq allows the isolation independent assessment of genetic accessibility of a microbe in community context. The DART vector system allows highly specific targeted DNA insertion
As a first step we developed Environmental Transformation Sequencing (ET-Seq), which allows us to determine if a microbe in a mixed community is susceptible to a DNA delivery and integration. Next, we developed the DNA-editing all-in-one RNA-guided CRISPR-Cas transposase (DART) system that enables organism- and locus- specific DNA integration. We have used these technologies individually and in combination to demonstrate enrichment of targeted species, confer novel metabolic traits, and measure gene fitness of bacteria within a community context in a lab setting. By developing and combining two new experimental tools (ET-Seq and DART) we have been able to specifically modify the DNA of individual bacteria while they exist in a community as well as track these modifications as the community grows.
Thus, we can begin to understand which bacteria contribute more to beneficial (i.e. promoting plant growth) vs. detrimental (i.e. releasing methane into the atmosphere) community collective behaviors. In the future, this technology may also allow us to directly modify specific bacteria in communities to do completely new and beneficial functions (e.g. design communities that efficiently eat plastic waste).
To read more about the metagenomics work at the Innovative Genomics Institute click here https://innovativegenomics.org/news/metagenomics-101-spencer-diamond/
Jennifer A. Doudna
Innovative Genomics Institute
University of California, Berkeley
doudna@berkeley.edu
Jillian F. Banfield
Innovative Genomics Institute
University of California, Berkeley
jbanfield@berkeley.edu