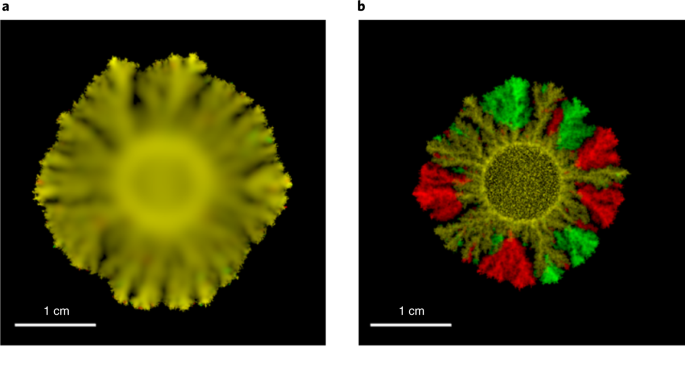
Similarly to the way computer simulations are used to forecast the weather, COMETS simulates how microbial communities and their surrounding environment change of over time, as a consequence of metabolic activity. In COMETS, metabolic networks of individual species are used to predict growth and the flow of molecules across the membrane. This is complemented by equations that describe how biomass and metabolites propagate in space, enabling complex virtual experiments with multiple species.
The newest version of COMETS, for which a recent paper provides a hands-on guide, includes several new capabilities, including extracellular enzymes, photosynthesis under day/night cycles, and evolutionary processes.
The Impact
Microbial communities have a major influence on our planet and on human life, often through their metabolic activity. In addition to directly contributing to biogeochemical cycles, microbes can modulate plant growth and biomass degradation. It is also increasingly useful to think of microbial communities as metabolic factories, capable of performing useful tasks better than individual, engineered microorganisms. Given their complexity, microbial communities cannot be understood and predicted without the help of computational models. We anticipate that comparisons between COMETS predictions and experimental data will help us test our understanding of microbial ecosystems and facilitate rational design of consortia with desired properties.
Summary
Genome-scale modeling of metabolism, utilizing the method of flux balance analysis, has become a standard systems biology tool for modeling cellular physiology and growth. Extensions of this approach are emerging as a valuable avenue for predicting, understanding and designing microbial communities. The software platform that we developed as a collaborative effort, Computation of microbial ecosystems in time and space (COMETS), extends dynamic flux balance analysis to generate simulations of multiple microbial species in molecularly complex and spatially structured environments. COMETS incorporates biophysical models of microbial biomass expansion upon growth, evolutionary dynamics, extracellular enzyme activity modules and other modeling capabilities. In addition to a command-line option, COMETS includes user-friendly Python and MATLAB interfaces compatible with the well-established COBRA models and methods, as well as comprehensive documentation and tutorials. In our recent publication in Nature Protocols we provide a detailed guideline for installing, testing and applying COMETS to different scenarios, generating simulations that take from a few minutes to several days to run, with broad applicability to microbial communities across biomes and scales. We provide several examples of COMETS applications to simulate the dynamics of complex microbial ecosystems, such as the soil microbiome and the day-night periodic variability in the metabolic activity of a marine autotroph.
Paper
Dukovski, I., Bajić, D., Chacón, J.M. et al. “A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS).” Nature Protocols (2021) doi.org/10.1038/s41596-021-00593-3
| Daniel Segre Boston University dsegre@bu.edu |