Four members of the m-CAFEs team recently participated in a virtual Research Slam promoting science related to the BioEPIC facility. Trent Northen, lead scientist, gave an overview presentation about the SFA. He was joined by m-CAFEs team members Manuel Villalobos, a postdoctoral researcher in the Glass Lab at UC Berkeley, and Avery Roberts, a graduate student in the Barrangou Lab at North Carolina State University. Manual and Avery presented their research and competed for the honor of Slam Champion. Cat Adams (m-CAFEs), also a postdoctoral researcher in the Glass Lab at UC Berkeley AND 2022 BioEPIC Research Slam Champion, was co-emcee for the event.
Results: Mingfei Chen (ENIGMA) took first place, Mo Ombadi (Watershed Function) second, and Avery Roberts (m-CAFEs) tied for honorable mention with Bradley Biggs (ENIGMA).
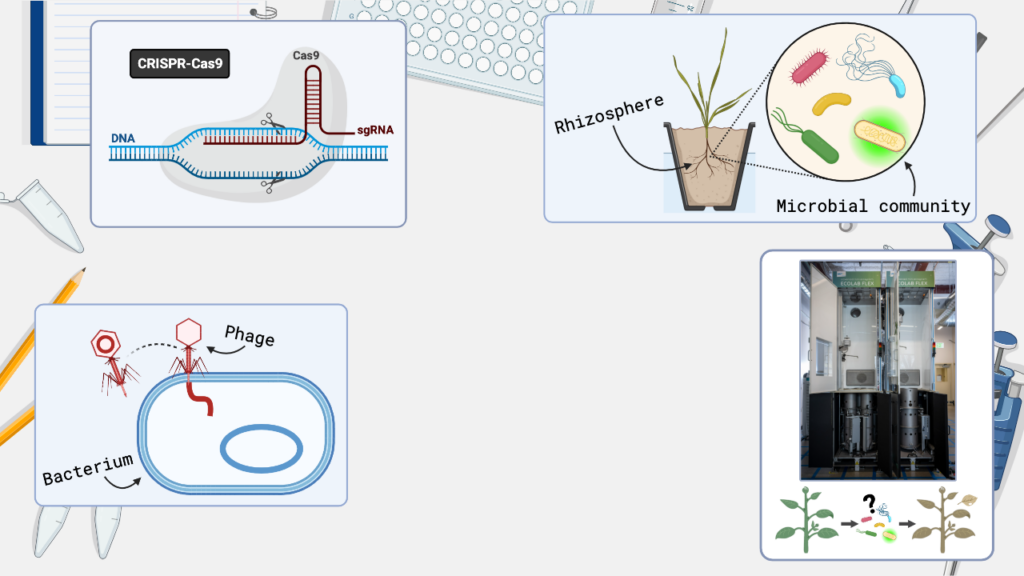
Avery’s research focuses on using genetically engineered viruses to make targeted changes to the DNA of bacteria within microbial communities. The ultimate goal of m-CAFEs is to apply these methods to the rhizosphere microbial community, helping us better understand microbial interactions and how these communities impact plant health. You can watch the SLAM event https://bioepic.lbl.gov/slam/ or read the text of Avery’s talk below.
“A CRISPR Crossover: Harnessing Anti-Phage Defenses for Targeted Editing of Microbial Communities”
2023 BioEPIC Virtual Research Slam Honorable Mention

So, bacteriophages, or phages, are viruses that infect and kill bacteria. Bacteria, in turn, defend themselves from phages using CRISPR-Cas systems, or CRISPR for short, which can locate and destroy invading phage DNA. CRISPR has been repurposed as a powerful gene editing tool and, in our research, we’ve reprogrammed CRISPR systems to engineer the DNA of phages, rather than destroy it.
Because phages have a natural talent for getting their DNA into bacterial cells, we’re using phages as a delivery system to deliver genes for CRISPR tools into bacteria. Once the engineered phage delivers its DNA, the bacteria produce the CRISPR machinery, which goes on to make precise changes, or edits, to the bacterial DNA. Currently, we’re engineering phages to deliver a specialized CRISPR system that can insert chunks of DNA at locations of our choosing within a bacterial genome. This technology will allow us to insert new genes or turn off genes of interest, or even tag bacteria with a fluorescent reporter, like this green fluorescent protein, for visualization within their community context.
With the successful development of these complex editing strategies, we’ll soon be taking the next step towards editing defined rhizosphere microbial communities created by the m-CAFEs team. This transition will enable us to use the advanced EcoPOD growth chambers, which are housed in the new BioEPIC building and designed for contained ecosystems and controlled environmental conditions. The EcoPOD provides a way for us to investigate how plants and microbes react when we alter the DNA of bacteria living within the rhizosphere by using our phage-based techniques. For example, in the EcoPOD we can use engineered phages to deliver CRISPR machinery that knocks out a critical gene in a rhizosphere microbe, and then measure the resulting plant health response.
Overall, we’re developing flexible phage-based tools for targeted gene editing in microbial communities to deepen our understanding of microbial interactions, and we’re working towards applying these techniques to the rhizosphere to open new possibilities to improve plant growth and health.