Soil microorganisms interact with plant roots in a variety of different ways. For example, microorganisms may carry out harmful activities such as infection, or sequestration of nutrients required for plant survival. In other cases, plant growth-promoting bacteria (PGPB) carry out beneficial activities such as nitrogen fixation, or protection from environmental stress (ex. from pathogens or drought). Microorganisms can also “communicate” with the plants via the production or destruction of chemical signals including plant hormones such as auxins. These signals and the microbial community members that modulate them can thus regulate root growth via chemical communication. Here, scientists led by Jeff Dangl at the University of North Carolina, Chapel Hill used molecular and genetic tools to uncover the mechanisms by which one bacterial genus (Variovorax) protects thale cress (A. thaliana) from auxin induced root growth inhibition.
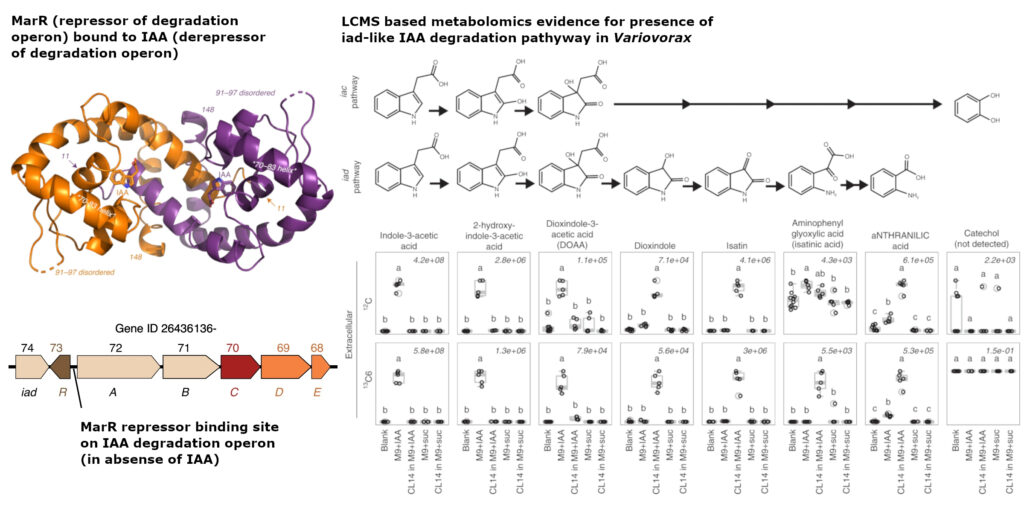
Using m-CAFEs metabolomics capabilities, the Northen lab at Lawrence Berkeley National Laboratory, identified the production of key intermediates specific to the iad-like degradation pathway of IAA in Variovorax, including dioxindole, isatin, isatinic acid along with iad-like specific final end product, anthranilic acid.
Plant roots and the surrounding soil are colonized by complex communities of soil microbes. Our understanding of how these communities and their individual members impact plant growth is very limited. Uncovering the genetic and molecular mechanisms by which bacteria impact root growth, has broad implications for improved agricultural practices. Use of newly identified plant growth-promoting bacteria or their chemical signals could be used to enhance crop productivity, reduce the use of fertilizers, and help protect crops against drought or pathogens.
Auxins are plant hormones that influence growth and development processes such as cell elongation, stem and root growth, and flowering. Some soil microorganisms produce the auxin, indole-3-acetic acid (IAA) which inhibits root growth in thale cress (Arabidopsis thaliana), a model flowering plant. Variovorax sp. have been demonstrated to revert this IAA induced root growth inhibition. In this study, scientists uncovered the molecular and genetic mechanism by which Variovorax achieves this, resulting in normal A. thaliana root growth, even in the presence of added IAA or IAA producing bacteria. Through phylogenetic analysis and characterization of representative IAA-degrading strains from diverse bacterial genera they found that only strains containing a particular type of auxin-degrading operons interfere with auxin signalling in a complex synthetic community context. Members of the m-CAFEs team characterized the IAA degradation products using liquid chromatography-mass spectrometry based metabolomics analysis of plant and bacterial cultures. Together, this work provides new mechanistic insights into bacterial modulation auxins in the plant microbiome.
Publication: Conway, J. M.; Walton, W. G.; Salas-González, I.; Law, T. F.; Lindberg, C. A.; Crook, L. E.; Kosina, S. M.; Fitzpatrick, C.R.; Lietzan, A. D.; Northen, T. R.; Jones, C. D.; Finkel, O. M.; Redinbo, M. R.; Dangl, J. L. (2022) Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome. Nature Microbiology 2022, 1–17. [DOI]:10.1038/s41564-022-01244-3 OSTI:1894181
Contact
Trent R. Northen
Lawrence Berkeley National Laboratory
Senior Scientist – Environmental Genomics and Systems Biology Division of Berkeley Lab
Deputy Director – Environmental Genomics and Systems Biology Division of Berkeley Lab
Laboratory Research Manager – m-CAFEs SFA Program,
Director of Biotechnology – ENIGMA SFA Program,
Director of High-Throughput Biochemistry – JBEI,
and Metabolomics Program Lead – the DOE JGI
trnorthen@lbl.gov
Office phone: (510) 495-8505; Cell phone: (858) 201-8998
Funding
This work was supported by NSF, the Howard Hughes Medical Institute and by the m-CAFEs Microbial Community Analysis & Functional Evaluation in Soils Science Focus Area.