Combining network analyses with metagenomics uncovers microbial interactions and a vitamin super-producer
m-CAFEs researchers at University of California, Berkeley demonstrated a new method for identifying bacteria that are interacting and determining the nature of the interaction. They predicted and confirmed experimentally that a Variovorax bacterium in a bioreactor community was producing significant amounts of vitamin B1 for other community members, yielding new insight into the relationships that structure these systems.
Credit: Image courtesy of Hessler
DOI:https://doi.org/10.1038/s41467-023-40360-4
The Science
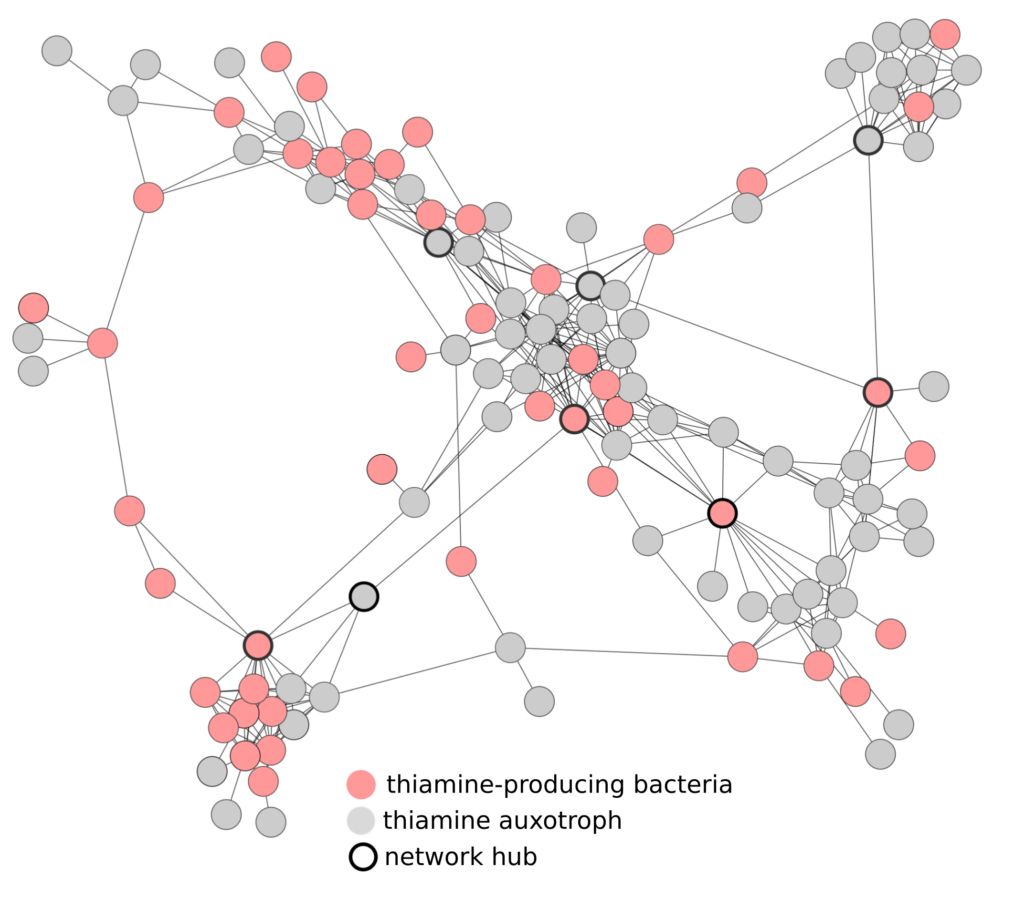
Bacteria seldom live alone, instead forming complex communities that interact with each other in cooperative and antagonistic ways. Microbiologists use many methods to unravel these interactions, including building networks from correlation data. However, this computational approach is typically done without laboratory-based functional validation.
Similarly, many studies use metagenomics, the study of genetic information for a community of different organisms, to predict microbial interactions, but these predictions are seldom tested experimentally. In this study, researchers integrated metagenomics with network analysis constructed from abundance correlation data to predict vitamin sharing in microbial communities. They then tested their prediction by isolating the bacterium of interest, Variovorax, and confirming its ability to produce large amounts of vitamin B1 (thiamine).
The Impact
The results of the study not only confirm the specific prediction about thiamine production but also show that combining network analyses and metagenomics is a powerful way to identify interactions within microbial communities. Understanding these types of interactions could ultimately lead to interventions and advancements in various fields where microbiome communities have a significant impact, including human health, agriculture, and environmental sciences.
Summary
Researchers developed a method to identify microorganisms in mixed communities with high vitamin-sharing potential. By analyzing correlations between organisms and examining their genomes using metagenomic data, they predicted which organisms are crucial for providing vitamins to other co-occurring members. To validate their prediction, they cultivated the bacterium predicted to be involved in thiamine production and found it produced significantly more thiamine than other known producers. The researchers hypothesize that this bacterium, Variovorax, may be producing thiamine to support the growth of surrounding bacteria. In return, Variovorax likely receives other vitamins and amino acids from these organisms. Remarkably, some of the bacteria predicted to rely on Variovorax for thiamine are rare and challenging to study in the lab. This study provides valuable insights into the ecology and physiology of these elusive bacteria, shedding light on their role and dependencies in microbial communities.
These findings have significant implications for understanding the interactions and dynamics within microbial communities. By identifying key players in vitamin biosynthesis and unraveling mutualistic relationships, researchers can gain insights into the intricate mechanisms that drive the structuring of microbial communities.
Read the full publication in Nature.
Contact
Tomas Hessler
University of California, Berkeley
tomas.hessler@berkeley.edu

Jillian F. Banfield
Innovative Genomics Institute
University of California, Berkeley
jbanfield@berkeley.edu
Publications
Hessler, T., Huddy, R.J., Sachdeva, R., Lei, S., Harrison, S.T., Diamond, S. and Banfield, J.F., 2023. Vitamin interdependencies predicted by metagenomics-informed network analyses and validated in microbial community microcosms. Nature Communications, 14(1), p.4768.
[DOI:https://doi.org/10.1038/s41467-023-40360-4] OSTI:1995510