The Science
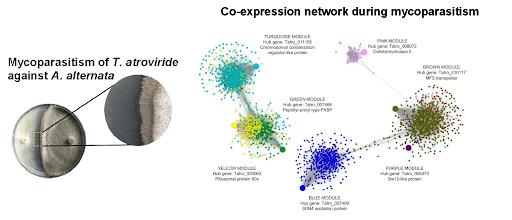
Fungi in the Trichomaderma genus, which are commonly present in soils, are known to engage in mycoparasitism, or active antagonism of other fungi. The fungi targeted in these interactions are phytopathogenic (plant-harming) fungi, which cause significant economic losses in agriculture. Although there has been extensive research about the mechanisms involved in the mycoparasitic capability of Trichoderma species, there are still unsolved questions related to how Trichoderma regulates recognition, attack, and defense mechanisms during interaction with a fungal host.
In this work, m-CAFEs researchers explored the participation of the Trichoderma atroviride RNA interference (RNAi) machinery, or the biological components involved in mRNA degradation induced by complementary sequences of small interfering RNA (siRNA), in the interaction with six phytopathogenic fungi of economic importance. They report that the Argonaute and Dicer components of the RNAi machinery and the small RNAs they process are essential for gene regulation during mycoparasitism by Trichoderma atroviride.
The Impact
There is a growing demand for plant disease control methods that don’t rely on chemical pesticides in order to mitigate environmental pollution, resistance, and associated health risks. Utilizing Trichoderma species in agriculture for fungal disease management presents an alternative approach that circumvents these issues without the use of chemical fungicides. Hence, comprehending the biocontrol mechanisms employed by Trichoderma species to antagonize other fungi is imperative. This work helped identify key genes involved in mycoparasitism, which can guide future efforts to enhance these strains and implement them in agricultural production.
Summary
Researchers utilized over ten mutants in the RNAi machinery, evaluating their interaction with six different pathogens. Using RNA-seq, an approach for measuring RNA expression levels, researchers found that RNAi appears to play an essential role in regulating multiple aspects of Trichoderma-host mycoparasitism interactions, including carbohydrate metabolism, cell recognition, signaling, and communication, and cell wall degradation. Beyond identifying Argonaute and Dicer as key genes in fungal biocontrol with the potential for future molecular improvement, their results reveal a fine-tuned regulation in T. atroviride dependent on siRNAs that is essential during the biocontrol of phytopathogenic fungi, showing a greater complexity of this process than previously established. Morover, these findings serve as a roadmap for identifying new Trichoderma strains with enhanced efficacy in fungal pest biocontrol.
Contact
José Manuel Villalobos Escobedo Plant and Microbial Biology Department, The University of California, Berkeley Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Lab jose.villalobos@berkeley.edu

Alfredo Herrera-Estrella
Laboratorio Nacional de Genómica para la Biodiversidad-Unidad de Genómica Avanzada, Cinvestav Campus Guanajuato, Irapuato, Guanajuato, Mexico alfredo.herrera@cinvestav.mx
Funding
This material by m-CAFEs Microbial Community Analysis & Functional Evaluation in Soils (m-CAFEs@lbl.gov), an SFA led by Lawrence Berkeley National Laboratory is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231.
Publications
Enriquez-Felix, E. E., Pérez-Salazar, C., Rico-Ruiz, J. G., Calheiros de Carvalho, A., Cruz-Morales, P., Villalobos-Escobedo, J. M., & Herrera-Estrella, A. (2024). Argonaute and Dicer are essential for communication between Trichoderma atroviride and fungal hosts during mycoparasitism. Microbiology Spectrum, 12(4), e03165-23. PMID: 38441469 PMCID: PMC10986496 DOI: 10.1128/spectrum.03165-23